Difference between revisions of "Mapping to ontologies for multiple gene sets (workflow)"
From BioUML platform
m (Protected "Mapping to ontologies for multiple gene sets (workflow)": Autogenerated page ([edit=sysop] (indefinite))) |
(GeneXplain -> geneXplain) |
||
| Line 2: | Line 2: | ||
:Mapping to ontologies for multiple gene sets | :Mapping to ontologies for multiple gene sets | ||
;Provider | ;Provider | ||
| − | :[[ | + | :[[geneXplain GmbH]] |
== Workflow overview == | == Workflow overview == | ||
[[File:Mapping-to-ontologies-for-multiple-gene-sets-workflow-overview.png|400px]] | [[File:Mapping-to-ontologies-for-multiple-gene-sets-workflow-overview.png|400px]] | ||
Revision as of 15:47, 18 April 2013
- Workflow title
- Mapping to ontologies for multiple gene sets
- Provider
- geneXplain GmbH
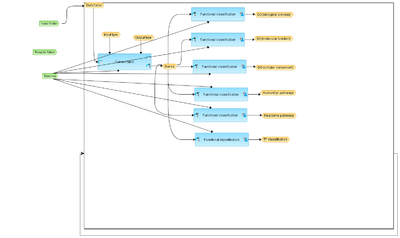
Workflow overview
Description
This workflow is designed to classify a multiple set of genes by enrichment analysis using GO, Reactome, HumanCyc and TF classification databases. Gene sets should be located in one folder.
Parameters
- Input folder
- Folder to get input tables from
- Species
- Results folder
- Folder to store results (will be created if not exists yet)