Difference between revisions of "Peaks to matrices (workflow)"
From BioUML platform
(Automatic synchronization with BioUML) |
(Automatic synchronization with BioUML) |
||
| (One intermediate revision by one user not shown) | |||
| Line 12: | Line 12: | ||
:BED file containing peaks | :BED file containing peaks | ||
;Sequences source | ;Sequences source | ||
| + | :Select Ensembl version which corresponds to your input data | ||
;Output folder | ;Output folder | ||
:Folder to store output | :Folder to store output | ||
Latest revision as of 17:23, 16 May 2013
- Workflow title
- Peaks to matrices
- Provider
- Autosome.Ru
[edit] Workflow overview
[edit] Description
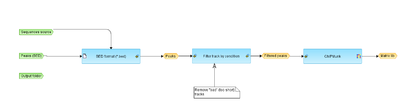
This workflow allows you to upload BED file which contains peaks produced by some peak finder, remove too short peaks and run ChIPMunk analysis to predict the motifs.
[edit] Parameters
- Peaks (BED)
- BED file containing peaks
- Sequences source
- Select Ensembl version which corresponds to your input data
- Output folder
- Folder to store output