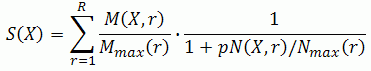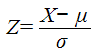Difference between revisions of "Regulator search (analysis)"
(Automatic synchronization with BioUML) |
(Automatic synchronization with BioUML) |
||
| (2 intermediate revisions by one user not shown) | |||
| Line 32: | Line 32: | ||
* '''Score cutoff''' – Molecules with Score lower than specified will be excluded from the result | * '''Score cutoff''' – Molecules with Score lower than specified will be excluded from the result | ||
* '''Search collection''' – Collection containing reactions | * '''Search collection''' – Collection containing reactions | ||
| + | * '''Custom search collection''' – Path to the custom search collection | ||
| + | * '''Relation sign''' – Consider only specified type of relation chain between molecules. | ||
* '''Species''' – Species to which analysis should be confined | * '''Species''' – Species to which analysis should be confined | ||
* '''Calculate FDR''' – If true, analysis will calculate False Discovery Rate | * '''Calculate FDR''' – If true, analysis will calculate False Discovery Rate | ||
| Line 37: | Line 39: | ||
* '''Z-score cutoff''' – Molecules with Z-score lower than specified will be excluded from the result | * '''Z-score cutoff''' – Molecules with Z-score lower than specified will be excluded from the result | ||
* '''Penalty''' (expert) – Penalty value for false positives | * '''Penalty''' (expert) – Penalty value for false positives | ||
| − | * ''' | + | * '''Decorators''' – Decorators |
| − | + | ||
| − | + | ||
* '''Normalize multi-forms''' (expert) – Normalize weights of multiple forms | * '''Normalize multi-forms''' (expert) – Normalize weights of multiple forms | ||
* '''Output name''' – Output name. | * '''Output name''' – Output name. | ||
| Line 53: | Line 53: | ||
Where: | Where: | ||
| − | * '''''R''''' | + | * '''''R''''' — Max radius (input parameter) |
| − | * '''''p''''' | + | * '''''p''''' — Penalty (input parameter) |
| − | * '''''N(X,r)''''' | + | * '''''N(X,r)''''' — total number of molecules reachable from key molecule X within the radius r. |
| − | * '''''N<sub>max</sub>(r)''''' | + | * '''''N<sub>max</sub>(r)''''' — maximal value of ''N(X,r)'' over all key molecules X found for this radius. |
| − | * '''''M(X,r)''''' | + | * '''''M(X,r)''''' — sum of ''w(X)'' for all hits reachable from key molecule X within the radius r, where ''w(X)'' — weight of hit X. It equals to ''w<sub>b</sub>(X)'' if “Normalize multi-forms” is unchecked. Otherwise it’s ''w<sub>b</sub>(X)/I(X)'', where ''I(X)'' is the number of multiforms of X in the input set (not total number of multiforms in the database). In both cases ''w<sub>b</sub>(X)'' is the base weight of hit X. It equals the corresponding value in “Weighting column” or 1 if “Weighting column” is not specified. |
| − | * '''''M<sub>max</sub>(r)''''' | + | * '''''M<sub>max</sub>(r)''''' — maximal value of ''M(X,r)'' over all key molecules X found for this radius. |
'''FDR''' Each individual drug target molecule gets a ''p''-value (FDR) assigned, which represents the probability to occupy the observed rank or higher ranks by random chance. It is estimated on-the-fly by random sampling. The ranking of the key nodes is defined by sorting them according to the Score above in descending order. It should be noted that the rank is defined by the ranks of the occurring scores, which means that more than one key node can share the same score value in some cases. Molecules which do not have any hits get assigned the last rank since the score is zero in this case. | '''FDR''' Each individual drug target molecule gets a ''p''-value (FDR) assigned, which represents the probability to occupy the observed rank or higher ranks by random chance. It is estimated on-the-fly by random sampling. The ranking of the key nodes is defined by sorting them according to the Score above in descending order. It should be noted that the rank is defined by the ranks of the occurring scores, which means that more than one key node can share the same score value in some cases. Molecules which do not have any hits get assigned the last rank since the score is zero in this case. | ||
| Line 79: | Line 79: | ||
==== Result columns ==== | ==== Result columns ==== | ||
| − | * '''ID''' | + | * '''ID''' — key molecule identifier in respective database |
| − | * '''Key molecule name''' | + | * '''Key molecule name''' — molecule title |
| − | * '''Reached from set''' | + | * '''Reached from set''' — number of molecules from input set, that were reached from key molecule within the distance given |
| − | * '''Reachable total''' | + | * '''Reachable total''' — total number of molecules, that can be reached from key molecule within the distance given |
| − | * '''Score''' | + | * '''Score''' — specificity score value calculated as described above |
| − | * '''FDR''' | + | * '''FDR''' — ''p''-value, which represents the probability to occupy the observed rank or higher ranks by random chance |
| − | * '''Z-Score''' | + | * '''Z-Score''' — z-score, according the equation above |
| − | * '''Hits''' | + | * '''Hits''' — identifiers of molecules from input set, that were reached from key molecule within the distance given |
| − | * '''Hits names''' | + | * '''Hits names''' — titles of molecules from input set, that were reached from key molecule within the distance given |
==== References: ==== | ==== References: ==== | ||
Latest revision as of 16:33, 12 March 2019
- Analysis title
 Regulator search
Regulator search
- Provider
- geneXplain GmbH
- Class
RegulatorKeyNodes- Plugin
- biouml.plugins.keynodes (Master regulator node analysis plugin)
Contents |
[edit] Drug target search analysis
Goal: to search for important molecules in signal transduction cascade.
Input: a set of genes / molecules to start analysis with. For instance, this can be a set of transcription factors, which may result from a promoter analysis, or a set of ligands / receptors that trigger a certain (set of) pathway(s).
Two separate analyses are available:
- Effector search for molecules downstream of the molecules in the input list.
- Regulator search for molecules upstream of the molecules in the input list.
[edit] Output:
A set of proteins or their encoding genes, which may play a key role in regulating (or being regulated by) a maximal number of start molecules.
[edit] Parameters:
- Molecules collection – Input the collection of molecules/genes
- Weighting column (expert) – Column to replace weights in search graph
- Limit input size (expert) – Limit size of input list
- Input size (expert) – Size of input list
- Max radius – Maximal search radius
- Score cutoff – Molecules with Score lower than specified will be excluded from the result
- Search collection – Collection containing reactions
- Custom search collection – Path to the custom search collection
- Relation sign – Consider only specified type of relation chain between molecules.
- Species – Species to which analysis should be confined
- Calculate FDR – If true, analysis will calculate False Discovery Rate
- FDR cutoff – Molecules with FDR higher than specified will be excluded from the result
- Z-score cutoff – Molecules with Z-score lower than specified will be excluded from the result
- Penalty (expert) – Penalty value for false positives
- Decorators – Decorators
- Normalize multi-forms (expert) – Normalize weights of multiple forms
- Output name – Output name.
[edit] Algorithm description
In drug target search analysis one searches for signaling molecules and corresponding networks that can transmit a signal to or receive a signal from several of input molecules within a certain limit of reaction steps. A search starts from each molecule of an input set Vx and constructs the shortest paths to all nodes V of the complete network within a given maximal path cost R (i.e, the sum of the costs of all edges in the shortest path from a vertex in Vx to a vertex in V should be smaller than or equal to R). The search can be conducted in reverse direction of the edges leading to input molecules (upstream) or in the same direction (downstream).
The Specificity score is calculated for every molecule found according to:
Where:
- R — Max radius (input parameter)
- p — Penalty (input parameter)
- N(X,r) — total number of molecules reachable from key molecule X within the radius r.
- Nmax(r) — maximal value of N(X,r) over all key molecules X found for this radius.
- M(X,r) — sum of w(X) for all hits reachable from key molecule X within the radius r, where w(X) — weight of hit X. It equals to wb(X) if “Normalize multi-forms” is unchecked. Otherwise it’s wb(X)/I(X), where I(X) is the number of multiforms of X in the input set (not total number of multiforms in the database). In both cases wb(X) is the base weight of hit X. It equals the corresponding value in “Weighting column” or 1 if “Weighting column” is not specified.
- Mmax(r) — maximal value of M(X,r) over all key molecules X found for this radius.
FDR Each individual drug target molecule gets a p-value (FDR) assigned, which represents the probability to occupy the observed rank or higher ranks by random chance. It is estimated on-the-fly by random sampling. The ranking of the key nodes is defined by sorting them according to the Score above in descending order. It should be noted that the rank is defined by the ranks of the occurring scores, which means that more than one key node can share the same score value in some cases. Molecules which do not have any hits get assigned the last rank since the score is zero in this case.
Z-Score
In addition to the FDR, each drug target molecule gets a Z-Score
which measures the deviation of the observed rank X of the key node from the expected rank μ in random case, divided by the standard deviation. In this formula, the rank above distribution is assumed to comply the normal distribution. Key nodes with Z greater than 1.0 are considered significant.
Context algorithm
For the purpose of incorporating additional contextual knowledge, e.g. a certain disease which we know to be related to the anticipated analysis, we implemented a method which encodes this additional context information as modified edge costs in the signaling network. The context information has to be provided as a second gene set (context genes). The idea is based on attracting the drug target molecule search (e.g. the underlying Dijkstra algorithm for shortest paths) towards context genes by decreasing the costs of those edges that are close to the context genes. It features two major aspects:
- Attraction ("gravity") of the shortest-paths towards context genes C
- Distribution of the attraction power to an extended surrounding area around C in order to prefer shortest paths close to context genes in case there is no path possible that goes through the context gene directly. ("gravity range").
[edit] Result columns
- ID — key molecule identifier in respective database
- Key molecule name — molecule title
- Reached from set — number of molecules from input set, that were reached from key molecule within the distance given
- Reachable total — total number of molecules, that can be reached from key molecule within the distance given
- Score — specificity score value calculated as described above
- FDR — p-value, which represents the probability to occupy the observed rank or higher ranks by random chance
- Z-Score — z-score, according the equation above
- Hits — identifiers of molecules from input set, that were reached from key molecule within the distance given
- Hits names — titles of molecules from input set, that were reached from key molecule within the distance given
[edit] References:
- Kel, A., Voss, N., Jauregui, R., Kel-Margoulis, O. and Wingender, E.: Beyond microarrays: Find key transcription factors controlling signal transduction pathways BMC Bioinformatics 7(Suppl. 2), S13 (2006).