Difference between revisions of "Convert identifiers for multiple gene sets (workflow)"
From BioUML platform
m (Protected "Convert identifiers for multiple gene sets (workflow)": Autogenerated page ([edit=sysop] (indefinite))) |
(Automatic synchronization with BioUML) |
||
| Line 6: | Line 6: | ||
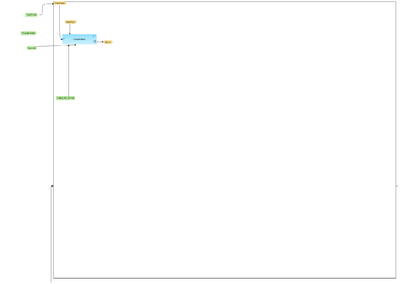
[[File:Convert-identifiers-for-multiple-gene-sets-workflow-overview.png|400px]] | [[File:Convert-identifiers-for-multiple-gene-sets-workflow-overview.png|400px]] | ||
== Description == | == Description == | ||
| − | This workflow is designed to convert several gene or protein tables into | + | This workflow is designed to convert several gene or protein tables into new gene tables with Ensembl IDs. The input is a folder containing several gene or protein tables, which needs to be converted. |
| − | The first input table is converted to a list with Ensembl IDs using ''Convert table'' analysis | + | The first input table is converted to a list with Ensembl IDs using ''Convert table'' analysis and same step is repeated for the next input table. Several cycles are performed, automatically corresponding to the number of total tables in the input folder. |
| − | The output is a new folder with | + | The output is a new folder with the new Ensembl tables. |
Latest revision as of 16:34, 12 March 2019
- Workflow title
- Convert identifiers for multiple gene sets
- Provider
- geneXplain GmbH
[edit] Workflow overview
[edit] Description
This workflow is designed to convert several gene or protein tables into new gene tables with Ensembl IDs. The input is a folder containing several gene or protein tables, which needs to be converted.
The first input table is converted to a list with Ensembl IDs using Convert table analysis and same step is repeated for the next input table. Several cycles are performed, automatically corresponding to the number of total tables in the input folder.
The output is a new folder with the new Ensembl tables.
[edit] Parameters
- Input folder
- Folder to get input tables from
- Species
- output_file_format
- Folder to store results (will be created if not exists yet)
- Results folder
- Folder to store results (will be created if not exists yet)