Difference between revisions of "GTRD Workflow"
Ivan Yevshin (Talk | contribs) |
|||
| (4 intermediate revisions by one user not shown) | |||
| Line 11: | Line 11: | ||
[https://groups.csail.mit.edu/cgs/gem/ GEM] <ref> Yuchun Guo, Shaun Mahony & David K Gifford. High Resolution Genome Wide Binding Event Finding and Motif Discovery Reveals Transcription Factor Spatial Binding Constraints. PLoS Computational Biology 8(8): e1002638. 2012.</ref> | [https://groups.csail.mit.edu/cgs/gem/ GEM] <ref> Yuchun Guo, Shaun Mahony & David K Gifford. High Resolution Genome Wide Binding Event Finding and Motif Discovery Reveals Transcription Factor Spatial Binding Constraints. PLoS Computational Biology 8(8): e1002638. 2012.</ref> | ||
and [http://www.bioconductor.org/packages/release/bioc/html/PICS.html PICS] <ref>Zhang X, Robertson G, Krzywinski M, Ning K, Droit A, Jones S and Gottardo R. “PICS: Probabilistic Inference for ChIP-seq.” Biometrics, 66. 2010.</ref>. | and [http://www.bioconductor.org/packages/release/bioc/html/PICS.html PICS] <ref>Zhang X, Robertson G, Krzywinski M, Ning K, Droit A, Jones S and Gottardo R. “PICS: Probabilistic Inference for ChIP-seq.” Biometrics, 66. 2010.</ref>. | ||
| + | |||
| + | Peaks computed for the same transcription factor and peak calling method, but different experiment conditions (e.g., cell line, treatment, etc.) were joined into clusters. | ||
| + | |||
| + | Clusters for the same TF revealed by different peak calling methods were joined into metaclusters. Metaclusters represent non-redundant set of transcription factor binding sites. | ||
| + | The clustering algorithm used in GTRD is described in the main GTRD paper <ref>Yevshin et al. GTRD: a database of transcription factor binding sites identified by ChIP-seq experiments. Nucleic Acids Res. 2017 Jan 4;45(D1):D61-D67.</ref> | ||
==Bowtie2== | ==Bowtie2== | ||
| Line 60: | Line 65: | ||
[[File: gtrd-workflow-pics-script.png]] | [[File: gtrd-workflow-pics-script.png]] | ||
| + | |||
| + | ==CPICS== | ||
| + | |||
| + | Starting from GTRD v17.04 we use [https://github.com/Biosoft-ru/cpics CPICS] as a replacement of PICS. CPICS is an efficient implementation of PICS ChIP-seq peak caller. | ||
| + | |||
| + | <code> | ||
| + | cpics -u $unmappable_regions_for_specie_and_readlen -c $control -g /srv/local-main/data/${species}.chrs $bam ${peaks}.bed | ||
| + | </code> | ||
| + | |||
| + | ==Clusters== | ||
| + | |||
| + | Peaks computed for the same transcription factor and peak calling method, but different experiment conditions (e.g., cell line, treatment, etc.) were joined into clusters. | ||
| + | The peaks with centers located 50 bp from each other or closer were merged into one cluster. | ||
| + | Depending on peak caller, we used different peak centers: for MACS we used the reported ‘summit’ column; GEM reports sites of unit length, and we used this coordinate as the peak center; for PICS and SISSRs, we used the geometric center of the peak. For each cluster, we found the center by computing the median of the peak centers. | ||
| + | |||
| + | The width of cluster reflect both the actual length of DNA interacting with the protein and uncertainty in the location of transcription factor binding site. | ||
| + | As an estimate of the actual length, the length of the position weight matrix (PWM) for the corresponding transcription factor available from the [http://hocomoco.autosome.ru HOCOMOCO database] was used. When such an estimate was not available, we used the fixed length of 20 bp. The uncertainty in the location of cluster was estimated from the variation of peak centers inside that cluster. Specifically, we have computed the standard deviation (SD) of peak centers inside each cluster and used 4*SD as the uncertainty factor. When a cluster was supported by only a single peak, the median of SD values from all other clusters was used instead of SD. The width of cluster was computed as a sum of actual length and uncertainty factor. | ||
| + | |||
| + | |||
| + | ==Metaclusters== | ||
| + | |||
| + | Clusters for the same transcription factor revealed by different peak calling methods were joined into metaclusters. Cluster centers located 50 bp from each other or closer were grouped, and one of them was selected based on priority of peak caller. Peak callers from high priority to low priority: GEM, PICS, MACS, SISSRs. Metaclusters supported by only one peak caller were filtered out. Metaclusters represent non-redundant set of transcription factor binding sites. | ||
==References== | ==References== | ||
<references /> | <references /> | ||
Latest revision as of 17:59, 29 November 2017
ChIP-seq experiment information were collected in semi-automated way from literature, GEO and ENCODE.
Raw ChIP-seq data in the form of fastq and SRA files were fetched from ENCODE and SRA databases.
Sequenced reads were aligned using Bowtie2 [1] aligner.
ChIP-seq peaks were called using 4 different methods: MACS [2] SISSRS [3] GEM [4] and PICS [5].
Peaks computed for the same transcription factor and peak calling method, but different experiment conditions (e.g., cell line, treatment, etc.) were joined into clusters.
Clusters for the same TF revealed by different peak calling methods were joined into metaclusters. Metaclusters represent non-redundant set of transcription factor binding sites. The clustering algorithm used in GTRD is described in the main GTRD paper [6]
Contents |
[edit] Bowtie2
We use bowtie2 version 2.2.3 for ChIP-seq read alignment to the reference genomes of human (GRCh38) and mouse (GRCm38).
Bowtie2 was run with following parameters:
bowtie2 -x $genome -U $fastq_files -p 8 --mm --seed 0
The resulting alignments were converted to bam files, then sorted and indexed using samtools version 1.0
[edit] MACS
MACS version 1.4.2 was used for peak calling with following parameters:
macs14 f BAM -g $species -n $peaks -t $alignment_bam
or if control experiment was available:
macs14 f BAM -g $species -n $peaks -t $alignment_bam -c $control_bam
[edit] SISSRS
SISSRS requires alignments in bed format, bam files were converted to bed files using bedtools version 2 by:
bamToBed -i $input_bam > $output_bed
Version 1.4 of SISSRS were used for peaks calling with following parameters:
sissrs.pl -i $alignment_bed -s 3000000000 -o $peaks.sissrs
or if control experiment was available:
sissrs.pl -i $alignment_bed -s 3000000000 -o $peaks.sissrs -b $control_bed
[edit] GEM
GEM version 2.5 was used with following parameters:
java -Xmx4G -XX:+UseSerialGC -jar /srv/local-main/tools/gem/gem.jar --d /srv/local-main/tools/gem/Read_Distribution_default.txt
--g /srv/local-main/tools/gem/$species.chrom.sizes --s 2000000000 --f SAM --t 1 --out $peaks --expt $bam
or if control experiment was available:
java -Xmx4G -XX:+UseSerialGC -jar /srv/local-main/tools/gem/gem.jar --d /srv/local-main/tools/gem/Read_Distribution_default.txt
--g /srv/local-main/tools/gem/$species.chrom.sizes --s 2000000000 --f SAM --t 1 --out $peaks --expt $bam --ctrl $control
For the large datasets -Xmx24G parameter was set.
[edit] PICS
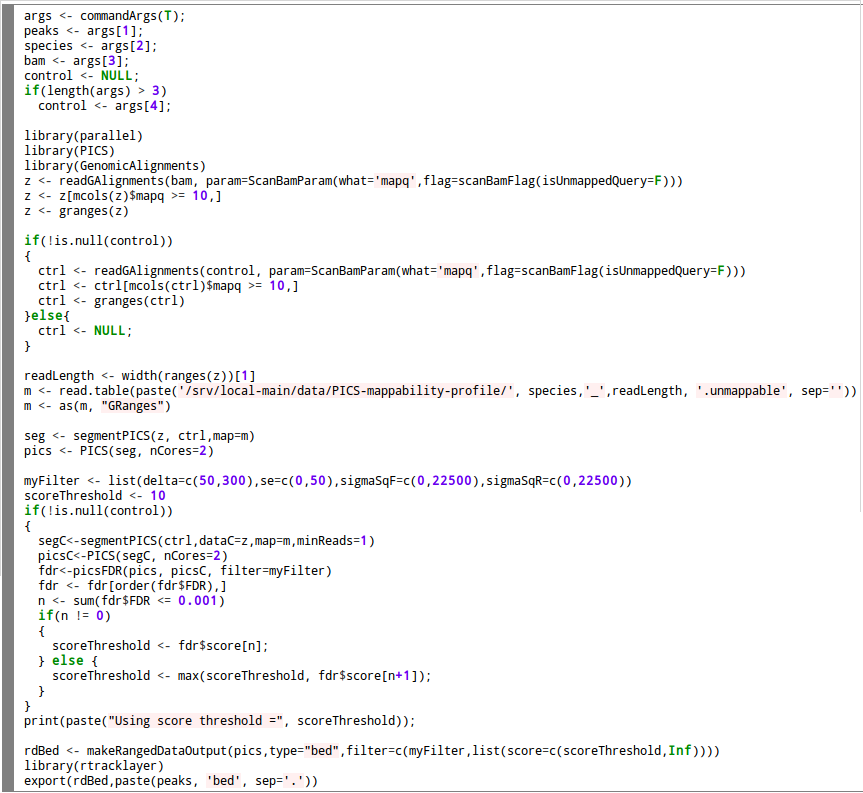
For peak calling with PICS method we use R version 3.2.0 and PICS version 2.12.0. We use the following custom R script:
[edit] CPICS
Starting from GTRD v17.04 we use CPICS as a replacement of PICS. CPICS is an efficient implementation of PICS ChIP-seq peak caller.
cpics -u $unmappable_regions_for_specie_and_readlen -c $control -g /srv/local-main/data/${species}.chrs $bam ${peaks}.bed
[edit] Clusters
Peaks computed for the same transcription factor and peak calling method, but different experiment conditions (e.g., cell line, treatment, etc.) were joined into clusters. The peaks with centers located 50 bp from each other or closer were merged into one cluster. Depending on peak caller, we used different peak centers: for MACS we used the reported ‘summit’ column; GEM reports sites of unit length, and we used this coordinate as the peak center; for PICS and SISSRs, we used the geometric center of the peak. For each cluster, we found the center by computing the median of the peak centers.
The width of cluster reflect both the actual length of DNA interacting with the protein and uncertainty in the location of transcription factor binding site. As an estimate of the actual length, the length of the position weight matrix (PWM) for the corresponding transcription factor available from the HOCOMOCO database was used. When such an estimate was not available, we used the fixed length of 20 bp. The uncertainty in the location of cluster was estimated from the variation of peak centers inside that cluster. Specifically, we have computed the standard deviation (SD) of peak centers inside each cluster and used 4*SD as the uncertainty factor. When a cluster was supported by only a single peak, the median of SD values from all other clusters was used instead of SD. The width of cluster was computed as a sum of actual length and uncertainty factor.
[edit] Metaclusters
Clusters for the same transcription factor revealed by different peak calling methods were joined into metaclusters. Cluster centers located 50 bp from each other or closer were grouped, and one of them was selected based on priority of peak caller. Peak callers from high priority to low priority: GEM, PICS, MACS, SISSRs. Metaclusters supported by only one peak caller were filtered out. Metaclusters represent non-redundant set of transcription factor binding sites.
[edit] References
- ↑ Langmead B, Salzberg S. Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012, 9:357-359.
- ↑ Zhang et al. Model-based Analysis of ChIP-Seq (MACS). Genome Biol (2008) vol. 9 (9) pp. R137
- ↑ Leelavati Narlikar, Raja Jothi. ChIP-Seq data analysis: identification of protein-DNA binding sites with SISSRs peak-finder. Methods in Molecular Biology, 802:305-22, 2012.
- ↑ Yuchun Guo, Shaun Mahony & David K Gifford. High Resolution Genome Wide Binding Event Finding and Motif Discovery Reveals Transcription Factor Spatial Binding Constraints. PLoS Computational Biology 8(8): e1002638. 2012.
- ↑ Zhang X, Robertson G, Krzywinski M, Ning K, Droit A, Jones S and Gottardo R. “PICS: Probabilistic Inference for ChIP-seq.” Biometrics, 66. 2010.
- ↑ Yevshin et al. GTRD: a database of transcription factor binding sites identified by ChIP-seq experiments. Nucleic Acids Res. 2017 Jan 4;45(D1):D61-D67.