Difference between revisions of "Genome Browser iframe"
Anna Ryabova (Talk | contribs) (Created page with "BioUML Genome Browser can be used as separate component. Genome Browser is developed as separate git project http://gitlab.dote.ru/isb/genome-browser-biouml It should be inst...") |
Anna Ryabova (Talk | contribs) |
||
| (5 intermediate revisions by one user not shown) | |||
| Line 1: | Line 1: | ||
| − | BioUML Genome Browser can be used as separate component. | + | '''BioUML Genome Browser''' can be used as separate component. |
| + | |||
Genome Browser is developed as separate git project http://gitlab.dote.ru/isb/genome-browser-biouml | Genome Browser is developed as separate git project http://gitlab.dote.ru/isb/genome-browser-biouml | ||
It should be installed as addition to BioUML server, since it uses server's javascripts, styles, images. | It should be installed as addition to BioUML server, since it uses server's javascripts, styles, images. | ||
| Line 5: | Line 6: | ||
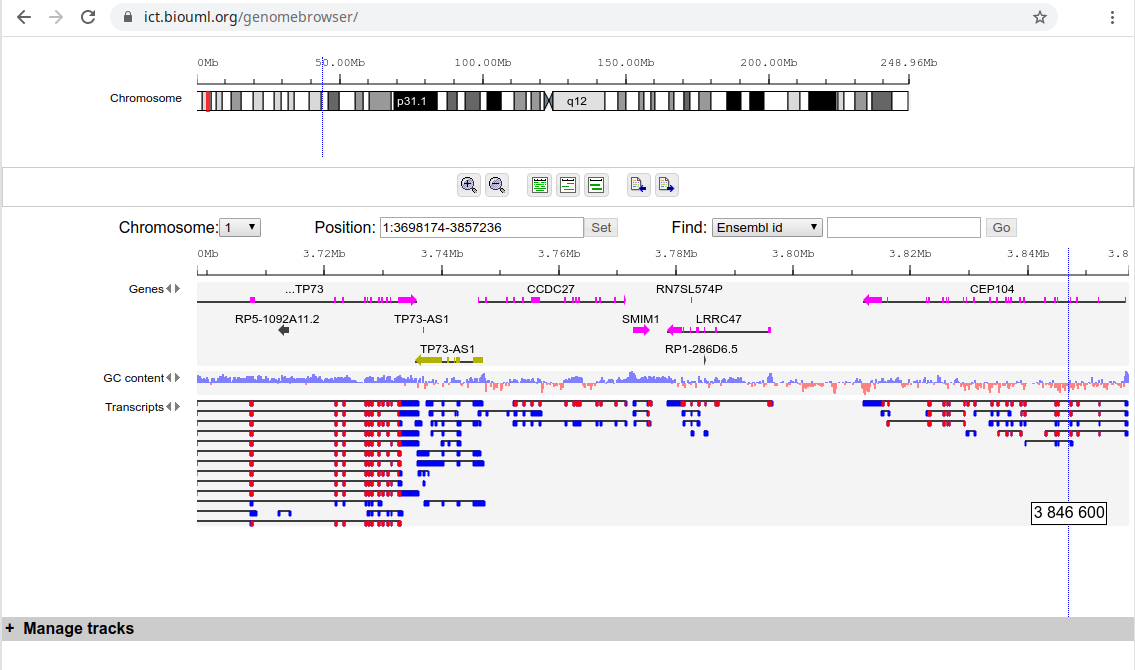
Here is the example of Genome Browser: https://ict.biouml.org/genomebrowser/ It uses BioUML server https://ict.biouml.org/bioumlweb | Here is the example of Genome Browser: https://ict.biouml.org/genomebrowser/ It uses BioUML server https://ict.biouml.org/bioumlweb | ||
| − | + | [[File:GB_browser.png]] | |
| − | + | ||
| − | The | + | The Genome Browser can be embed as iframe to any page as following:<br> |
| + | <iframe src="... page with Genome Browser..." title="BioUML Genome Browser" style="width:100%; height:100%;"></iframe> | ||
| − | + | The genome build, list of available tracks, start positions in genome and some other features can be configured for each installation of the Genome Browser in several ways:<br> | |
| − | + | ||
| − | 2. Settings can be passes as url parameter "json=". Javascript object with settings should be converted to url string, for example, via encodeURIComponent(JSON.stringify(json_object)) methods. | + | 1. Json file passed as url parameter "setup=..." to iframe or to a single page. The json config file is described [[Genome Browser json]]. It should be placed in the same domain, as Genome Browser page. To use files from another domain, Access-Control-Allow-Origin header should be send.<br> |
| − | https://ict.biouml.org/genomebrowser/?json=%7B%22sequencePath%22%3A%22databases%2FEnsemblMouse81_38%2FSequences%2Fchromosomes%20GRCm38%22%2C%22chromosome%22%3A%2210%22%2C%22from%22%3A1945654%2C%22to%22%3A4294028%2C%22chromosomeTrack%22%3A%22%22%2C%22tracks%22%3A%7B%22Genes%22%3A%22databases%2FEnsemblMouse81_38%2FTracks%2FGenes%22%2C%22Karyotype%22%3A%22databases%2FEnsemblMouse81_38%2FTracks%2FKaryotype%22%7D%2C%22initialTracks%22%3A%5B%22Genes%22%2C%22Karyotype%22%5D%7D | + | Example: https://ict.biouml.org/genomebrowser/?setup=ens85.json |
| + | <br><br> | ||
| + | 2. Settings can be passes as url parameter "json=". Javascript object with settings should be converted to url string, for example, via encodeURIComponent(JSON.stringify(json_object)) methods.<br> | ||
| + | Example: https://ict.biouml.org/genomebrowser/?json=%7B%22sequencePath%22%3A%22databases%2FEnsemblMouse81_38%2FSequences%2Fchromosomes%20GRCm38%22%2C%22chromosome%22%3A%2210%22%2C%22from%22%3A1945654%2C%22to%22%3A4294028%2C%22chromosomeTrack%22%3A%22%22%2C%22tracks%22%3A%7B%22Genes%22%3A%22databases%2FEnsemblMouse81_38%2FTracks%2FGenes%22%2C%22Karyotype%22%3A%22databases%2FEnsemblMouse81_38%2FTracks%2FKaryotype%22%7D%2C%22initialTracks%22%3A%5B%22Genes%22%2C%22Karyotype%22%5D%7D | ||
Latest revision as of 08:55, 4 February 2022
BioUML Genome Browser can be used as separate component.
Genome Browser is developed as separate git project http://gitlab.dote.ru/isb/genome-browser-biouml It should be installed as addition to BioUML server, since it uses server's javascripts, styles, images.
Here is the example of Genome Browser: https://ict.biouml.org/genomebrowser/ It uses BioUML server https://ict.biouml.org/bioumlweb
The Genome Browser can be embed as iframe to any page as following:
<iframe src="... page with Genome Browser..." title="BioUML Genome Browser" style="width:100%; height:100%;"></iframe>
The genome build, list of available tracks, start positions in genome and some other features can be configured for each installation of the Genome Browser in several ways:
1. Json file passed as url parameter "setup=..." to iframe or to a single page. The json config file is described Genome Browser json. It should be placed in the same domain, as Genome Browser page. To use files from another domain, Access-Control-Allow-Origin header should be send.
Example: https://ict.biouml.org/genomebrowser/?setup=ens85.json
2. Settings can be passes as url parameter "json=". Javascript object with settings should be converted to url string, for example, via encodeURIComponent(JSON.stringify(json_object)) methods.
Example: https://ict.biouml.org/genomebrowser/?json=%7B%22sequencePath%22%3A%22databases%2FEnsemblMouse81_38%2FSequences%2Fchromosomes%20GRCm38%22%2C%22chromosome%22%3A%2210%22%2C%22from%22%3A1945654%2C%22to%22%3A4294028%2C%22chromosomeTrack%22%3A%22%22%2C%22tracks%22%3A%7B%22Genes%22%3A%22databases%2FEnsemblMouse81_38%2FTracks%2FGenes%22%2C%22Karyotype%22%3A%22databases%2FEnsemblMouse81_38%2FTracks%2FKaryotype%22%7D%2C%22initialTracks%22%3A%5B%22Genes%22%2C%22Karyotype%22%5D%7D