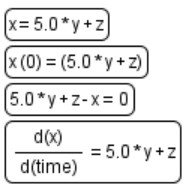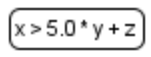Difference between revisions of "SBGN extension"
Ilya Kiselev (Talk | contribs) |
Ilya Kiselev (Talk | contribs) |
||
| (6 intermediate revisions by one user not shown) | |||
| Line 1: | Line 1: | ||
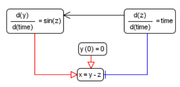
| + | [[File:sbgn_extension_example.png|thumb|Fig. 1. Example of relations between equations.]] | ||
| + | [[File:sbgn_extension_cvs_1.png|thumb|Fig. 2. Model of the human Cardiovascular system in BioUML.]] | ||
| + | [[File:sbgn_extension_cvs_2.png|thumb|Fig. 3. Model of the human Cardiovascular system in BioUML, concize form.]] | ||
BioUML fully supports and heavily utilizes SBML (Systems Biology Markup Language) as a standard for description of mathematical models with ODE and discrete events (https://sbml.org/). | BioUML fully supports and heavily utilizes SBML (Systems Biology Markup Language) as a standard for description of mathematical models with ODE and discrete events (https://sbml.org/). | ||
| Line 5: | Line 8: | ||
The most well known graphical notation in the systems biology is SBGN (Systems Biology Graphic Notation). It has three different versions, here we will concern only with Process description notation which describes the temporal courses of biochemical interactions in a network (https://sbgn.github.io/). It is compatible with SBML although it more relies on pathways comprising of entities and processes between them. Thus it does not cover every single object in SBML models. Particularly there is no visual representations for "mathematical" objects: equations, functions, events, etc. | The most well known graphical notation in the systems biology is SBGN (Systems Biology Graphic Notation). It has three different versions, here we will concern only with Process description notation which describes the temporal courses of biochemical interactions in a network (https://sbgn.github.io/). It is compatible with SBML although it more relies on pathways comprising of entities and processes between them. Thus it does not cover every single object in SBML models. Particularly there is no visual representations for "mathematical" objects: equations, functions, events, etc. | ||
| − | In BioUML we fully support SBGN Process Description notation, however we decided to extend notation we use in BioUML to fully represent SBML models. | + | In BioUML we fully support SBGN Process Description notation, however we decided to extend notation we use in BioUML to fully represent SBML models. Additional motivation for that is the fact that there are many mathematical objects of biological systems (in particular - physiological model) that are not represented by a network of biochemical reactions but rather contain a set of ODE equations. |
| + | Example of the model using only mathematical elements ([[Short term model of the heart output regulation]]) is represented on Fig.1. | ||
| + | More concise way is to omit equations right hand side is represented on Fig.2. | ||
| + | |||
| + | ===New elements added to SBGN notation in BioUML=== | ||
| + | |||
| + | <center> | ||
| + | {| class="wikitable" | ||
| + | !Name!!Notation!!Description | ||
| + | |- style="background-color: Lavender" | ||
| + | |Equations | ||
| + | |[[File:equations.png]] | ||
| + | |Mathematical equation in the model: <ul><li>assignment<li>initial asssignment<li>algebraic<li>ODE</ul> | ||
| + | |- | ||
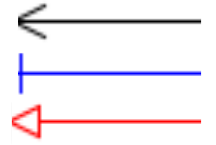
| + | |Relations | ||
| + | |[[File:sbgn_extension_relations.png]] | ||
| + | |Relations between equations are autogenerated and describes how variable calculated in one equations affect variable calculated in another equations. | ||
| + | there are three types: <ol><li>red - value increases (activation)<li>blue - value decreases (inhibition)<li>black - effect undefined</ol> | ||
| + | |- | ||
| + | |Events | ||
| + | |[[File:sbgn_extension_event.png]] | ||
| + | |Discrete event which describes instantaneous changes in the model variables when certain condition is met. E.g. change in drug intake after certain time point. | ||
| + | |- | ||
| + | |Function | ||
| + | |[[File:sbgn_extension_function.png]] | ||
| + | | Function takes argument values and calculates output. | ||
| + | |- | ||
| + | |Constraint | ||
| + | |[[File:sbgn_extension_constraint.png]] | ||
| + | |Constraint is a condition which is checked during the simulation. If the condition is violated then either error message is issued or simulation is stopped depending on simulator options. | ||
| + | |- | ||
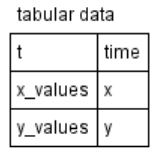
| + | |Tabular data | ||
| + | |[[File:sbgn_extension_tabular.png]] | ||
| + | |Tabular element refers to external table data collection and is used to calculate model variables based on data in said table. For example in this case table column t corresponds to model time, | ||
| + | table column x_values contains numerical data for model variable x. There are two ways to process table element: <ol><li>spline approximation<li>constant piecewise function</ol> | ||
| + | |- | ||
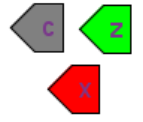
| + | |Ports | ||
| + | |[[File:sbgn_extension_ports.png]] | ||
| + | |In our notation ports corresponds to tags and terminals in SBGN. Color of the port denotes its type: <ol><li>gray - contact<li>green - inpu<li>red - output</ol> | ||
| + | |} | ||
Latest revision as of 20:02, 4 February 2022
BioUML fully supports and heavily utilizes SBML (Systems Biology Markup Language) as a standard for description of mathematical models with ODE and discrete events (https://sbml.org/). BioUML also utilizes Visual modeling as one of its core features. Thus each SBML model is represented visually in BioUML interface. Main component of visual modeling is formal graphic notation which defines how particular elements of the model should be represented visually.
The most well known graphical notation in the systems biology is SBGN (Systems Biology Graphic Notation). It has three different versions, here we will concern only with Process description notation which describes the temporal courses of biochemical interactions in a network (https://sbgn.github.io/). It is compatible with SBML although it more relies on pathways comprising of entities and processes between them. Thus it does not cover every single object in SBML models. Particularly there is no visual representations for "mathematical" objects: equations, functions, events, etc.
In BioUML we fully support SBGN Process Description notation, however we decided to extend notation we use in BioUML to fully represent SBML models. Additional motivation for that is the fact that there are many mathematical objects of biological systems (in particular - physiological model) that are not represented by a network of biochemical reactions but rather contain a set of ODE equations. Example of the model using only mathematical elements (Short term model of the heart output regulation) is represented on Fig.1. More concise way is to omit equations right hand side is represented on Fig.2.