Difference between revisions of "Flux balance analysis"
(→FBA via Analysis tab) |
|||
| Line 1: | Line 1: | ||
| + | [https://www.ncbi.nlm.nih.gov/labs/pmc/articles/PMC3108565/ Flux balance analysis] is a mathematical approach that enables to predict the flux distribution through a metabolic network on a genome-scale level. | ||
| + | |||
==FBA via Diagram== | ==FBA via Diagram== | ||
Revision as of 18:17, 14 March 2022
Flux balance analysis is a mathematical approach that enables to predict the flux distribution through a metabolic network on a genome-scale level.
Contents |
FBA via Diagram
FBA via Analysis tab
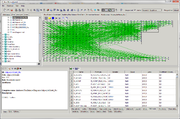
In the BioUML it is represented in the Flux Balance Constraint analysis. This page contains brief instruction of how this analysis should be used.
FBC analysis usage
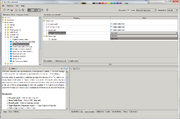
To start work with the Flux Balance Constraint analysis one needs a ![]() diagram and a special table with data about its fluxes (names, bounds, objective function coefficients, etc.). Analysis enables to select path to the
diagram and a special table with data about its fluxes (names, bounds, objective function coefficients, etc.). Analysis enables to select path to the ![]() diagram, to the data table and to the table with results. Also it has an expert mode where user can select type of the objective function, solver type (apache simplex solver and gurobi solver are available) and some solver's properties. If the
diagram, to the data table and to the table with results. Also it has an expert mode where user can select type of the objective function, solver type (apache simplex solver and gurobi solver are available) and some solver's properties. If the ![]() diagram contains fluxes properties written as fbc extension of the SBML "FBC table" tab can be used. Advantage of this tab usage is lack of necessity in creation table with data about fluxes: it will be created automatically using
diagram contains fluxes properties written as fbc extension of the SBML "FBC table" tab can be used. Advantage of this tab usage is lack of necessity in creation table with data about fluxes: it will be created automatically using ![]() diagram.
diagram.
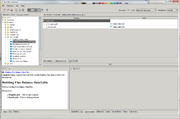
FBC data table creation
Table with flux balance constraint data can be created via Building Flux Balance Data Table analysis.
It is worth noting, that the Building Flux Balance Data Table analysis and "FBC table" tab work only with SBML L3v1 ![]() diagrams with special fbc package. Thus if the
diagrams with special fbc package. Thus if the ![]() diagram does not satisfy these conditions it should be transformed before using this analysis. For example, if the
diagram does not satisfy these conditions it should be transformed before using this analysis. For example, if the ![]() diagram does not use fbc package but contains all necessary information about fluxes, it should be firtsly preprocessed by Recon transformer.
diagram does not use fbc package but contains all necessary information about fluxes, it should be firtsly preprocessed by Recon transformer.