Difference between revisions of "Find 10 master regulators in networks (TRANSPATH(R)) (workflow)"
(Automatic synchronization with BioUML) |
m (Protected "Find 10 master regulators in networks (TRANSPATH(R)) (workflow)": Autogenerated page ([edit=sysop] (indefinite))) |
Latest revision as of 19:02, 13 February 2017
- Workflow title
- Find 10 master regulators in networks (TRANSPATH(R))
- Provider
- geneXplain GmbH
[edit] Workflow overview
[edit] Description
Variant of the workflow:
Visualization of top 10 master regular networks, instead of 3 in our "standard" workflows
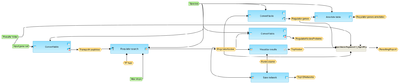
This workflow is designed to find master regulatory molecules upstream of an input list of genes. Input file is any gene or protein table.
At the first step, the input table is converted into a table with TRANSPATH® peptides.
This table is subjected to master regulator search in the TRANSPATH® network. For each potential master regulator, FDR, Score, and Z-score are calculated.
The results are filtered by Z_Score>1 and Score>0.2 to select statistically significant master regulators.
Further, the filtered regulatory molecules are converted to both Ensembl Gene IDs and a list of UniProt protein IDs.
The table with Ensembl Gene Ids is annotated with additional information, gene description and gene symbols.
Finally, the table with master regulatory molecules is sorted by Score and networks for the three top master regulators are visualized as diagrams in the hierarchical layout.
This workflow is available together with a valid TRANSPATH® license.
[edit] Parameters
- Input gene set
- Species
- Max steps
- Select max number of steps upstream
- Results folder