Analyze any DNA sequence for site enrichment (TRANSFAC(R)) (workflow)
- Workflow title
- Analyze any DNA sequence for site enrichment (TRANSFAC(R))
- Provider
- geneXplain GmbH
Workflow overview
Description
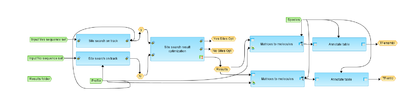
This workflow is designed to search for overrepresented transcription factor binding sites (TFBSs) in a collection of sequences (Input Yes sequence set) compared to a background collection of sequences. As input, any sequence collection from human, mouse or rat species can be submitted. Background sequences can be housekeeping gene promoters, sequences from a control experiment (e.g. media) or gene sequences, which are NOT induced/expressed. We recommend to use 3x more sequences in the background set as in the Yes set.
Sequences are analyzed for potential enriched cis-regulatory sites. Site search is done with the help of the TRANSFAC® library of positional weight matrices (PWMs), namely with the default profile (a matrix collection): vertebrate_non_redundant_minSUM.
At this step, frequencies of putative TFBSs are compared between Yes sequence set and a No sequence set to identify sites overrepresented in Yes set versus No set. The result of this step is a list of transcription factor binding sites, which are overrepresented in Yes set versus No set. Next, the list of PWMs is converted into a table of corresponding transcription factors and are annotated with additional information (gene description and gene symbols).
The output is a new folder with several tables, including a summary of the predicted TFBSs, a track of the Yes sequences and the identified enriched sites. As well as the final table with transcription factors, that are potentially regulating the genes in the Yes sequence set.
This workflow is available together with a valid TRANSFAC® license.
Parameters
- Input Yes sequence set
- Select Yes sequence set
- Input No sequence set
- Select No sequence set
- Species
- Profile
- Select Profile
- Results folder
- Select Results folder