Compute differentially expressed genes using Hypergeometric test (Affymetrix probes) (workflow)
- Workflow title
- Compute differentially expressed genes using Hypergeometric test (Affymetrix probes)
- Provider
- geneXplain GmbH
Workflow overview
Description
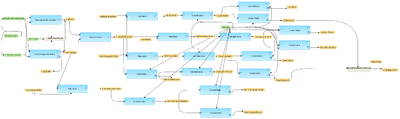
This workflow is designed to identify differentially expressed genes from an experiment data set compared to a control data set. It can be used even for the case with one data point for each the experiment and the control. Normalized data with Affymetrix probeset IDs can be submitted as input. Such normalized files are the output of the workflow normalization/Normalize Affymetrix experiment and control Normalize Affymetrix experiment and control.
In the first step, the up- and down-regulated probes are identified and log fold change values are calculated for all probes using the Fold Change calculation analysis. This workflow applies Hypergeometric analysis for the p-value calculation.
In addition the results are filtered by different conditions in parallel applying the Filter table method, to identify up-regulated and down-regulated Affymetrix probeset IDs. The filtering criteria are set as follows:
For up-regulated probes: LogFoldChange>0.5 and -log_P_value_>3.
For down- regulated probes: LogFoldChange<-0.5 and -log_P_value_<-3.
The resulting tables of up-regulated, down-regulated, and non-changed Affymetrix probeset IDs are converted into a gene table with the Convert table method and annotated with additional information (gene descriptions, gene symbols, and species) via Annotate table method.
A result folder is generated and automatically named corresponding to the experiment data set name. This resulting folder contains all generated tables.
Parameters
- Experiment normalized
- Control normalized
- Species
- Probe type
- Results folder