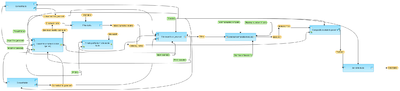
Identify enriched composite modules in promoters (GTRD) (workflow)
- Workflow title
- Identify enriched composite modules in promoters (GTRD)
- Provider
- geneXplain GmbH
Workflow overview
Description
This workflow is designed to find pairs of sites in the promoters of the input gene set. This workflow enables the identification of combinations of several enriched TFBSs in the promoters of the genes under study (Yes-set). The resulting composite module differentiates the Yes-set from a background set (No-set)
In the first part of the workflow, the enriched motifs are identified by the method Search for enriched TFBSs. The default profile used is the uniprobe profile from GTRD database. Motifs with an enrichment of >1.0 fold serve as a basis for constructing a specific profile, and this profile is used to run site search on the promoters of this input gene set .
In the second part of this workflow, composite modules are identified based on the enriched TFBSs.
This workflow identifies pairs of sites. By default, the minimum and maximum numbers of pairs are given as 2 and 8. You can change these parameters according to the number of pairs you aim to identify. The number of iterations of the genetic algorithm is 300 by default, and can be adapted as required.
The results folder consists of several folders and files including the enriched motifs table, composite modules table and Profile.
Note: This workflow may take more time depending on the size of the Yes and No sets and on the number of iterations. The recommended size of the Yes set is 150 genes maximum, and the recommended size of the No set is 300 genes maximum. The maximum recommended number of iterations is 300
"
Parameters
- Input Yes gene set
- Input No gene set
- Species
- Profile
- Minimal number of pairs
- Maximal number of pairs
- Number of iterations
- Start promoter
- End promoter
- Result folder