Mapping to GO ontologies and comparison for two gene sets (workflow)
- Workflow title
- Mapping to GO ontologies and comparison for two gene sets
- Provider
- geneXplain GmbH
Workflow overview
Description
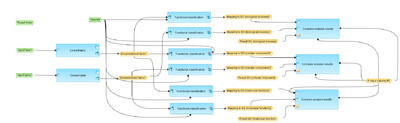
This workflow is designed to classify two input gene sets to several ontologies and to identify terms, hits for which are overrepresented in the input set and to compare the results of the functional classification.
The input file can be any gene or protein table. At the first step, the input table is converted into a table with Ensembl Gene IDs. This table with Ensembl Gene Ids is subjected to functional classification, which is done in parallel by the following ontologies: GO biological processes, GO cellular components, GO molecular functions, Reactome pathways, HumanCyc pathways, TF classification.
For each ontological term several parameters are calculated, including expected number of hits, actual number of hits, p-value, as well as hit names and the link to the corresponding ontological term.
Each ontological term is subjected to ‘Compare Analysis method’.
Output folder contains separate folder for each GO ontology, Analysis comparison plot and analysis comparison table. The Analysis comparison annot table lists identifiers, annotation of identifiers, P-values for the first input set of genes and P-values for the second input set of genes (-log). The column Difference shows the absolute difference between two P-values.
The columns Difference P-value and Difference FDR show the statistical significance of the absolute difference and upon sorting by one of these two columns on top you can see those ontology terms that are statistically most significantly different between two input gene sets.
The comparison reveals GO terms that show different enrichment across the two input gene lists.
Parameters
- InputTable1
- InputTable2
- Species
- Result folder