Difference between revisions of "BioUML web edition"
Ivan Yevshin (Talk | contribs) |
|||
| (11 intermediate revisions by 5 users not shown) | |||
| Line 1: | Line 1: | ||
| − | |||
| − | |||
[[File:BioUML web edition screenshot.png|thumb|BioUML web edition]] | [[File:BioUML web edition screenshot.png|thumb|BioUML web edition]] | ||
| − | BioUML web edition is a "thin" client for [[BioUML server]] (you just need to start your web browser and go to the dedicated web address) that provides most of the functionality of [[BioUML workbench]]. It uses [ | + | BioUML web edition is a "thin" client for [[BioUML server]] (you just need to start your web browser and go to the dedicated web address) that provides most of the functionality of [[BioUML workbench]]. It uses [[wikipedia:AJAX|AJAX]] and [[wikipedia:Canvas element|HTML5 <canvas>]] technology for visual modeling and interactive data editing. |
The current version is {{BioUML current version}}. | The current version is {{BioUML current version}}. | ||
| − | BioUML web edition is | + | A free BioUML web edition version is available at http://ie.biouml.org/bioumlweb. |
| + | |||
| + | The full commercial platform is available as geneXplain platform at http://platform.genexplain.com | ||
| + | |||
| + | The geneXplain high performance server available at https://155.253.6.35 | ||
==Minimum system requirements== | ==Minimum system requirements== | ||
| + | The user's computer must meet the following minimum requirements: | ||
'''Hardware''' | '''Hardware''' | ||
| Line 21: | Line 24: | ||
* Windows XP Service Pack 2+ or later, Mac OS X 10.6 or later, latest stable release of Linux or Android | * Windows XP Service Pack 2+ or later, Mac OS X 10.6 or later, latest stable release of Linux or Android | ||
| − | * latest web browser version (Google Chrome or | + | * latest web browser version (Google Chrome or Mozilla Firefox preferred) |
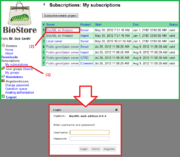
| + | [[File:Access via subscription centre.png|thumb|Accessing the login page of the web edition via the BioStore subscription centre]] | ||
==Starting work== | ==Starting work== | ||
| − | |||
Once you have completed the subscription procedure successfully on the [[BioStore]] website, the selected server can be accessed by | Once you have completed the subscription procedure successfully on the [[BioStore]] website, the selected server can be accessed by | ||
| − | |||
* clicking on '''Login to the server to start working''' link in the successful registration notification right away, | * clicking on '''Login to the server to start working''' link in the successful registration notification right away, | ||
* via the subscription management centre on the [[BioStore]] website: go to ''Menu > Subscriptions > My subscriptions'', choose the required server from the list of your valid subscriptions and click on the link in the Server column, | * via the subscription management centre on the [[BioStore]] website: go to ''Menu > Subscriptions > My subscriptions'', choose the required server from the list of your valid subscriptions and click on the link in the Server column, | ||
| Line 40: | Line 42: | ||
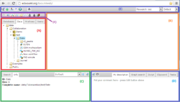
As you have logged in to the server, the BioUML workbench will open in your browser window, where the following can be found (see the figure): | As you have logged in to the server, the BioUML workbench will open in your browser window, where the following can be found (see the figure): | ||
| − | A | + | A — the tabbed '''[[repository pane]]''', where you find a collection of '''Databases''', the uploaded '''Data''' files and the available '''Analyses''' methods (under the corresponding tabs), organized in a hierarchical tree structure. Here you can also see a list of '''Users''' involved with the current project. |
| + | |||
| + | B — the '''[[document pane]]''' (aka main pane or workspace), which is the part of the window where contents of projects and databases are viewed and handled. There can be as many tabs in the pane as there are diagrams, workflows, tables etc. opened in the workbench at the moment. | ||
| − | + | C — the '''[[information box]]''' containing tabs for the '''property inspector''', which displays information about the data file or analysis method selected in the navigation pane above, and the '''text search engine'''. | |
| − | + | D — the '''[[viewpart|viewparts area]]''', providing a number of options under the different tabs in a context-dependent manner. | |
| − | + | E — the '''general control panel''', the topmost bar showing a context-dependent set of icons for the available operations. | |
| − | + | F — the '''[[repository pane toolbar]]''' containing a set of icons for the available operations depending on the item selected in the tree structure of the navigation pane. | |
| − | + | [[Category:Software]] | |
Latest revision as of 14:12, 1 December 2014
BioUML web edition is a "thin" client for BioUML server (you just need to start your web browser and go to the dedicated web address) that provides most of the functionality of BioUML workbench. It uses AJAX and HTML5 <canvas> technology for visual modeling and interactive data editing.
The current version is 2023.3.
A free BioUML web edition version is available at http://ie.biouml.org/bioumlweb.
The full commercial platform is available as geneXplain platform at http://platform.genexplain.com
The geneXplain high performance server available at https://155.253.6.35
[edit] Minimum system requirements
The user's computer must meet the following minimum requirements:
Hardware
- Intel Pentium IV or later, Athlon 64 or later
- 512 MB RAM
- 100 MB free hard disk drive space
Software
- Windows XP Service Pack 2+ or later, Mac OS X 10.6 or later, latest stable release of Linux or Android
- latest web browser version (Google Chrome or Mozilla Firefox preferred)
[edit] Starting work
Once you have completed the subscription procedure successfully on the BioStore website, the selected server can be accessed by
- clicking on Login to the server to start working link in the successful registration notification right away,
- via the subscription management centre on the BioStore website: go to Menu > Subscriptions > My subscriptions, choose the required server from the list of your valid subscriptions and click on the link in the Server column,
- entering the URI directly in the address bar of your web browser or activating the appropriate browser bookmark that you saved from the previous working session.
Whatever the starting point, you will be redirected to the login page of the web edition and prompted for the username and password. Complete the fields with the data required (your registered email for the username and the password you were sent by email) and press the Login button - the web interface of the workbench will open in the same browser window as pre-designed for this type of subscription.
[edit] User interface
As you have logged in to the server, the BioUML workbench will open in your browser window, where the following can be found (see the figure):
A — the tabbed repository pane, where you find a collection of Databases, the uploaded Data files and the available Analyses methods (under the corresponding tabs), organized in a hierarchical tree structure. Here you can also see a list of Users involved with the current project.
B — the document pane (aka main pane or workspace), which is the part of the window where contents of projects and databases are viewed and handled. There can be as many tabs in the pane as there are diagrams, workflows, tables etc. opened in the workbench at the moment.
C — the information box containing tabs for the property inspector, which displays information about the data file or analysis method selected in the navigation pane above, and the text search engine.
D — the viewparts area, providing a number of options under the different tabs in a context-dependent manner.
E — the general control panel, the topmost bar showing a context-dependent set of icons for the available operations.
F — the repository pane toolbar containing a set of icons for the available operations depending on the item selected in the tree structure of the navigation pane.